A New Method for Identifying Ancient DNA
A team of researchers have developed a new software called PyDamage to provide an automated approach for ancient DNA damage estimation and authentication of de novo DNA assemblies
De novo DNA assembly is a method for reconstructing genomes from a large quantity of DNA fragments, without knowing the original genomic source of those fragments. The method is increasingly used to obtain genomic data from ancient microbial communities, including from feces and dental calculus, to investigate the diversity and functionality of past microbiomes.
To obtain meaningful results, researchers must be able to distinguish ancient DNA from modern DNA contaminants. However, the tools that currently exist to identify ancient DNA are designed to find damage within the DNA of a single known reference sequence, such as the human genome for example, but not for the massive numbers of sequences from multiple unknown sources typical of de novo assembly.
To address this, researchers from the Max Planck Institute for the Science of Human History developed PyDamage, an automated approach for estimating damage and authenticating de novo assemblies of ancient DNA, now published in the journal PeerJ.
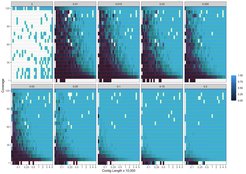
The software uses a likelihood ratio to rapidly assess damage patterns for numerous reference sequences in parallel, allowing researchers to profile the damage within a segment of DNA. In other words, the software is performing a statistical estimation of the odds of a DNA sequence to be contaminated. This allows researchers to distinguish between truly ancient stretches of DNA and stretches that derive from modern contamination.
In a new study, the team of researchers tested PyDamage on both simulated and empirical ancient DNA data from archaeological feces and demonstrated its ability to reliably and automatically identify DNA segments bearing damage characteristic of ancient DNA. PyDamage was most reliable in predicting damage for DNA segments with higher coverage, longer lengths, and higher damage, as expected, and importantly provides a statistical framework for damage evaluation that aids researchers in study design and sequencing plans.
“DNA de novo assembly of ancient DNA remains challenging, but now we have a tool to tell us if what we assembled is likely to be ancient,” says Maxime Borry, one of the authors of the study.
As the fields of microbiology and evolutionary biology increasingly turn to the archaeological record to investigate the dynamic evolutionary history of microbial communities, tools that reliably assemble and authenticate ancient metagenomic DNA are vital for understanding the microbial communities around and within us. Coupled with ancient DNA de novo assembly, PyDamage opens up new doors to explore and understand the functional diversity of ancient metagenomes.
